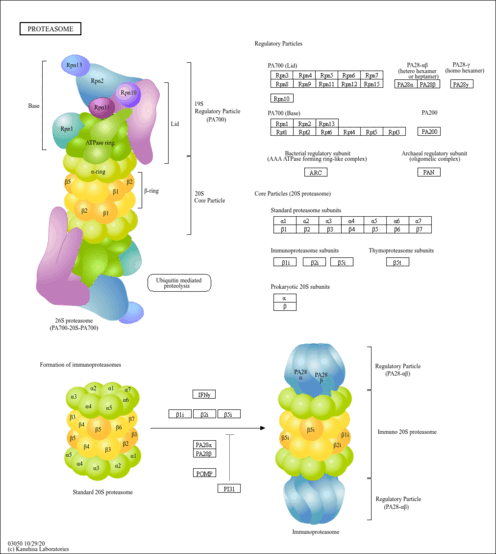
Proteasome, The Proteasome
Proteasomes are multicatalytic complexes containing a cylindrical 20S core which is composed of four heteroheptameric rings Harshbarger et al 2015The two inner β-rings contain the six proteolytic sites where substrates are cleaved. The reagents are added to test samples containing proteasome enzyme that cleaves the substrates releasing luciferin which is consumed by luciferase producing glow-type.

A Practical Review Of Proteasome Pharmacology Pharmacological Reviews
In eukaryotic cells degradation of most intracellular proteins is realized by proteasomes.
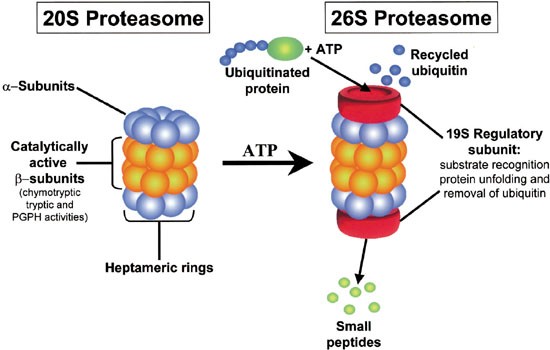
Proteasome. It is similar to the normal proteasome but has three new catalytic subunits swapped into the core. Proteasomes form the major non-lysosomal ATP-dependent protein degradation systems that targets intracellular polyubiquitinated proteins derived from self-structures or foreign structures for proteolytic degradation which includes the removal of misfolded and immature proteins and the production of peptides for presentation by MHC class I molecules. As noted above the proteasome core is actually a cylinder like hsp60.
A protein degradation machine within the cell that can digest a variety of proteins into short polypeptides and amino acids. It requires ATP to work. To supply amino acids for fresh protein synthesis.
The outer membrane of the proteasome admits only proteins carrying a ubiquitin molecule which detaches before entering the proteasome and is reused. Eucaryotice proteasomes are very similar yet they employ many more different subunits to construct the chamber. The proteasome is one of the major degradation machineries in eukaryotic cells.
There are two major intracellular organelles to digest damaged or unneeded proteins. Before a protein is degraded it is first flagged for destruction by the ubi. Protein degradation is essential to the cell.
We will focus on archaebacterial proteasomes because they are simpler. 20S Proteasome Chamber in Archaebacteria. To remove excess enzymes.
Medical Definition of proteasome. Proteasome - an organelle of intracellular digestion. Proteins to be destroyed are recognized by proteasomes because of the presence of ubiquitin conjugated to the targeted proteins lysine residue.
One of these subunits cleaves the chain next hydrophobic amino acids creating peptides that anchor particularly well to MHC. Prōtē-ă-sōm Cytoplasmic organelle composed of a cylindric core particle bound by two regulatory particles at each end responsible for degrading endogenous proteins. Lets consider the structure of the proteasome core.
Each ring has caspase-like C-L trypsin-like T-L and chymotrypsin-like ChT-L. However very little has been known about the mechanisms that control and execute the destruction of the proteasome. A hollow cylindrical cellular structure that is a complex of proteases involved in the selective degradation of intracellular proteins and especially proteins marked for proteolysis by ubiquitin Tagged proteins are then fed into the proteasome a barrel-shaped multi-protein complex that chops proteins down.
Proteins to be destroyed are recognized by proteasomes because of the presence of ubiquitin conjugated to the targeted proteins lysine residue. It is hollow and has openings at both ends to. The proteasome is a multisubunit enzyme complex that plays a central role in the regulation of proteins that control cell-cycle progression and apoptosis and has therefore become an important target for anticancer therapy.
The proteasome is itself made up of proteins. In Avram Hershkoaccompanies the protein to a proteasome which is essentially a structure of powerful enzymes that break the protein into its component amino acids. A specialized proteasome termed the immunoproteasome is induced during the immune response and performs the cleavages.
The substrates for proteolysis are selected by the fact that the gate to the proteolytic chamber of the proteasome is usually closed and only proteins carrying a special label can get into it. Proteasome prōtē-ă-sōm Cytoplasmic organelle composed of a cylindric core particle bound by two regulatory particles at each end responsible for degrading endogenous proteins. It terminates the existence of thousands of short-lived damaged misfolded or otherwise obsolete proteins and plays pivotal roles in protein quality control and other vital processes in the cell.
The 26S proteasome is a 25MDa multiprotein complex found in all eukaryotic cells. To remove transcription factors gene action that are no longer needed. Modulating Proteasome Activity by Changing Proteasome Composition.
AM 114 a derivative of boronic chalcone is a potent small-molecule inhibitor of the proteasome that inhibits the chymotrypsin-like activity of the 20S proteasome with a value of 50 inhibition concentration IC50 of approximately 1 uM resulting in a significant accumulation of ubiquitinated p53 and other cellular.

The Proteasome A Novel Target For Cancer Chemotherapy Leukemia

Protein Degradation By The Ubiquitin Proteasome Pathway In Normal And Disease States American Society Of Nephrology

Purified Bioactive Proteasomes R D Systems

Biomolecules Free Full Text The Ubiquitin Proteasome System In Immune Cells

Infographic Proteasome Basics The Scientist Magazine
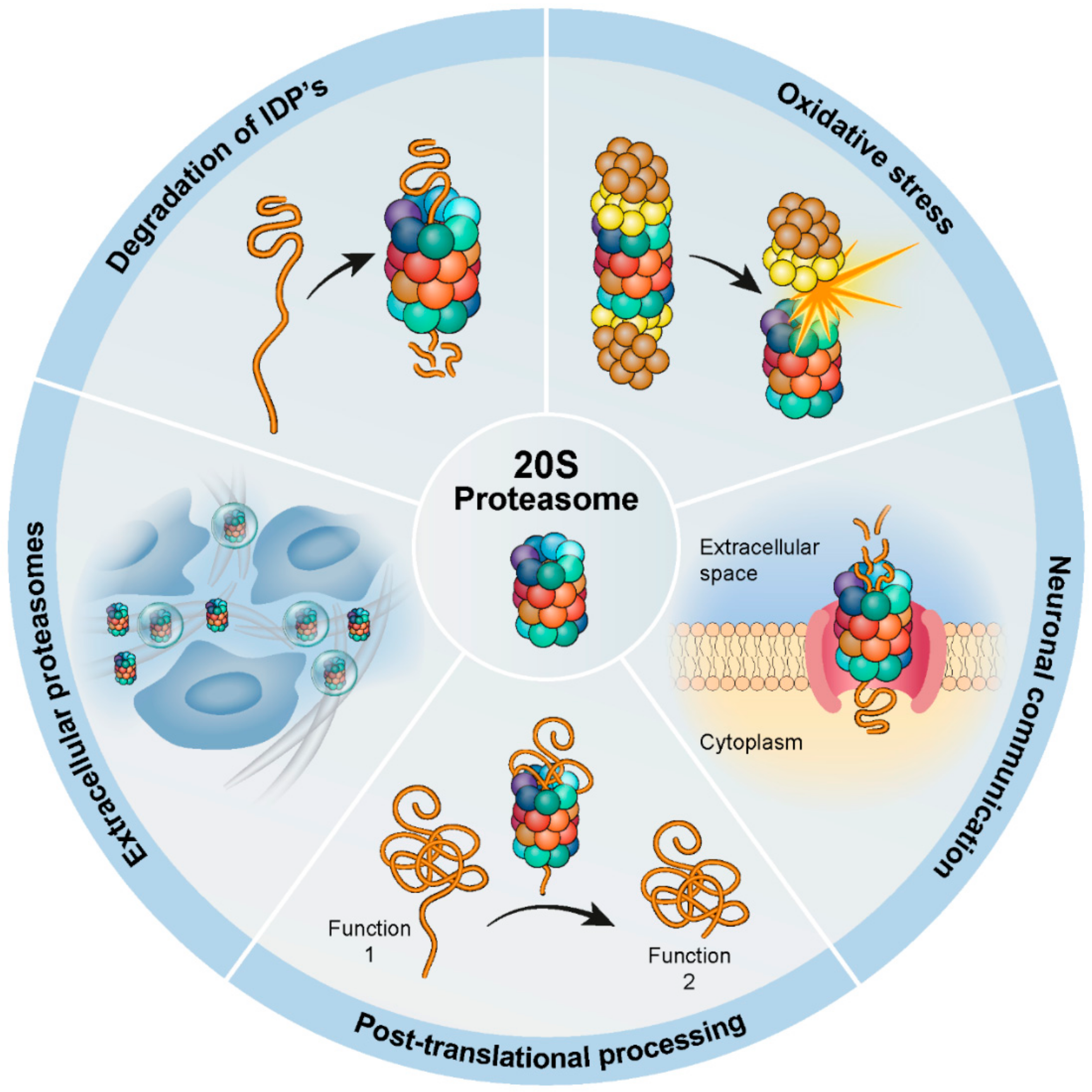
Biomolecules Free Full Text The Contribution Of The 20s Proteasome To Proteostasis Html
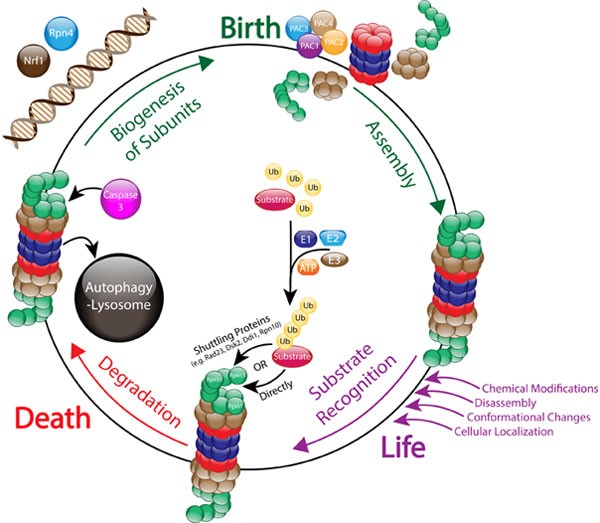
The Life Cycle Of The 26s Proteasome From Birth Through Regulation And Function And Onto Its Death Cell Research
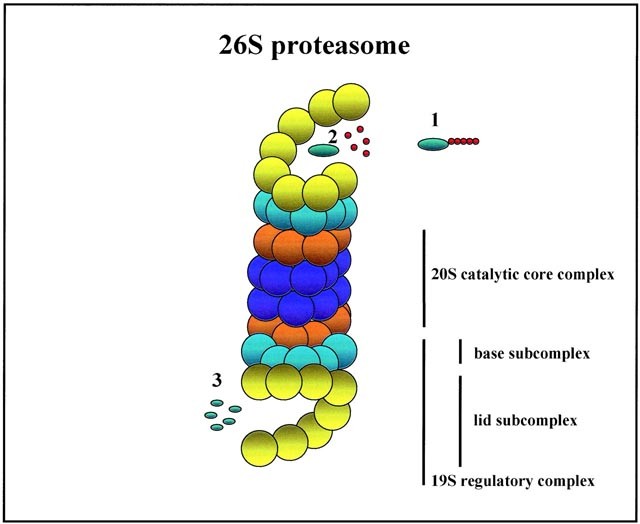
Role And Function Of The 26s Proteasome In Proliferation And Apoptosis Laboratory Investigation
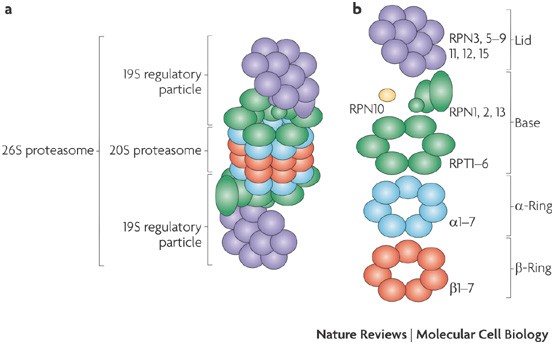
Molecular Mechanisms Of Proteasome Assembly Nature Reviews Molecular Cell Biology

Priming The Proteasome To Protect Against Proteotoxicity Trends In Molecular Medicine

How The Proteasome Is Degraded Pnas
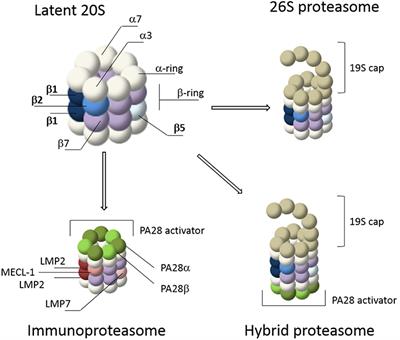
Frontiers Visualizing Proteasome Activity And Intracellular Localization Using Fluorescent Proteins And Activity Based Probes Molecular Biosciences

Pdb 101 Molecule Of The Month Proteasome
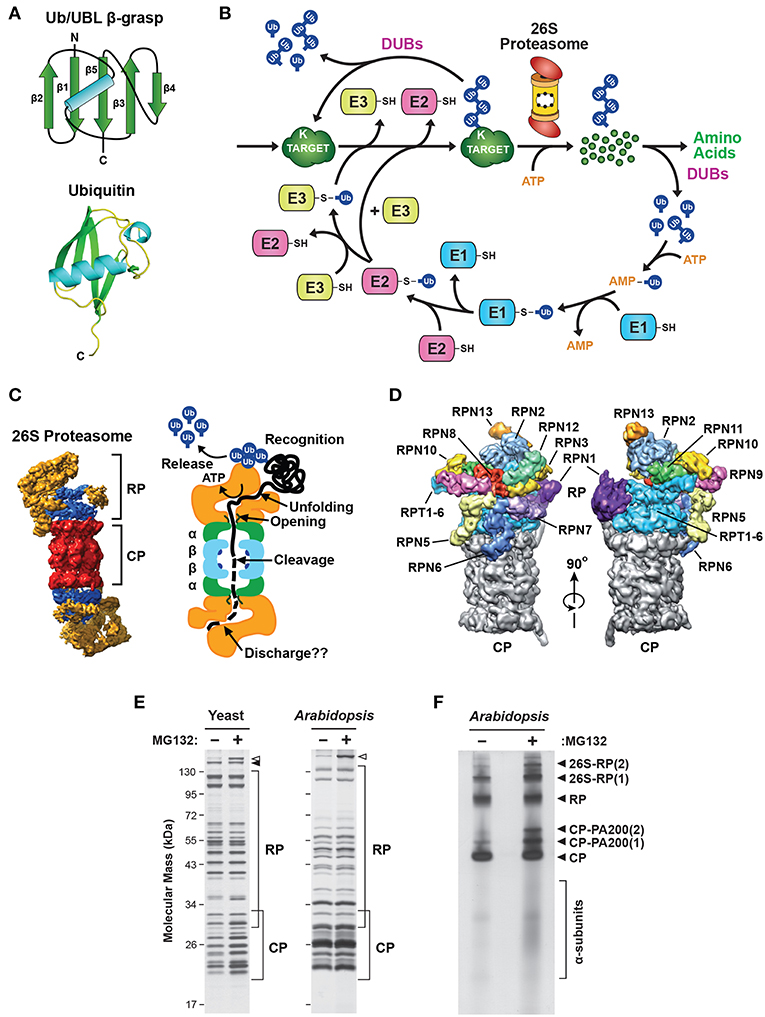
Frontiers Dynamic Regulation Of The 26s Proteasome From Synthesis To Degradation Molecular Biosciences

Schematic Representation Of Proteasome 26s Proteasome 26s Named Due Download Scientific Diagram